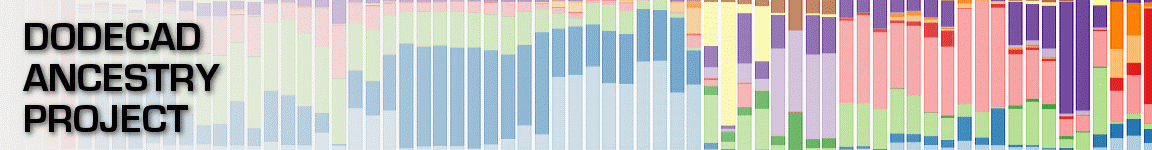
I took the 25 different inferred clusters from my recent
ChromoPainter analysis, and calculated their normalized median components in terms of the
K12b calculator. This is a quite useful exercise, since it can show in what sense clusters are different from each other.
Here are two ways in which you may use this correspondence.
1. Different clusters of a single population
For example, the Turks with partial Balkan ancestry
tend to belong to pop10, whereas those of Anatolian ancestry to pop13, and those from northeastern Anatolia to pop22. If we compare the admixture proportions of these three groups, we notice e.g.,
- An excess of Atlantic_Baltic and North_European in pop10
- An excess of Caucasus in pop22
Or, there is a group of 5 Iranians that belong to pop12, whereas the overwhelming majority of Iranians and Kurds belong to pop21. Strikingly, pop12 differs from all other populations in having substantial levels of East_African and Sub_Saharan. So, it seems that fineSTRUCTURE was able to infer that some Iranian individuals had this feature in common. These individuals were already evident in the Iranian population portrait (right), but fineSTRUCTURE was able to group them even though there were no African populations in the ChromoPainter analysis; presumably, the software was able to detect that these individuals shared a set of chunks that were quite different than is the norm for the Balkan/West Asian area.
2. Related clusters
fineSTRUCTURE grouped the different populations in a
tree structure. For example, it grouped pop18, the "North Balkan" cluster with pop23, the "Bulgarian-Romanian" one.
Looking at the admixture proportions, we can tell that the two clusters do indeed seem quite similar, but there are some differences, e.g., an excess of North_European in pop18, and an excess of Caucasus in pop23. This makes sense given the geographical origin of individuals belonging to the two clusters.